Our clinical reports are generated through Djerba, which houses our 47 clinical workflows. The report is generated for individual patients based on the results of sequencing and are accredited for clinical use. The document contains the assay results, interpretation of said results, and any treatment that might be recommended based on the results. Please find below more information about our accredited assays, pipelines and clinical reports.
Validated Pipelines for Accredited Assays
Whole Genome and Transcriptome Case Package
Our validated tumor-normal whole genome and transcriptome (WGTS) clinical case package includes 80X tumor, 30X normal whole genome and 100M paired end whole transcriptome in one integrated report. A lower coverage version of this package is also available when high tumor purity input is provided (40X tumor, 30X normal WGS, 100M PE WTS).
The report details oncogenic copy number variants (CNVs), somatic single nucleotide variants (SNVs), insertion-deletions (indels), structural variants (SVs), loss of heterozygosity (LOH), fusions, inferred cancer cell content (purity), genome-wide ploidy and fusions with OncoKb annotation and evidence tiers. All clinical reports are reviewed by board-certified geneticists (ACMG/CCMG).
Target Sequencing Clinical Case Package
Our validated tumor-normal targeted (TAR) clinical case package includes an ultra-deep sequenced tumor and normal pair. The specific sequencing depth will be determined by the needs of the project. The targeted sequencing pipeline utilizes Unique Molecular Identifier (UMIs), which are added to sequencing libraries before any PCR amplification steps. UMIs enable the accurate bioinformatic identification of PCR duplicates. This allows precise genomic variant detection, especially for low frequency mutations. The report details oncogenic somatic single nucleotide variants (SNVs) and insertion-deletions (indels). All clinical reports are reviewed by board-certified geneticists (ACMG/CCMG).
Shallow Whole Genome Package
Our validated tumor-normal shallow whole genome (SW) clinical case package includes an ultra-low depth sequenced tumor and normal pair. The SW pipeline estimates tumor fractions and predicts large-scale copy number variation (CNV).
The Pathway from FASTQ to Clinical Reporting
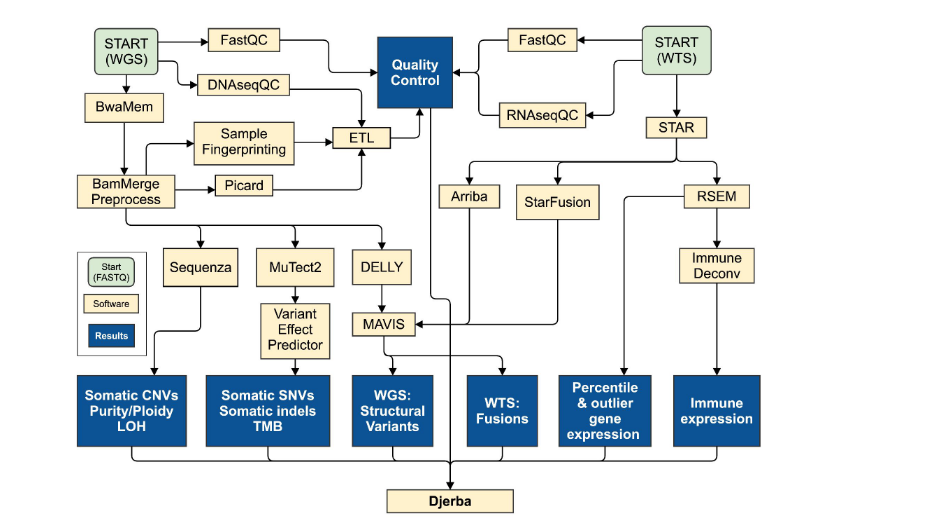
Clinical Report Breakdown

Case and Assay Overview
This section includes key information provided by the sample submitter allowing for the sample to be processed correctly through the laboratory and informatics pipelines. Information my include the primary cancer diagnosis, the site of biopsy/surgery, and the unique IDs assigned to track the submission.
Treatment Options
This section outlines any treatments and/or investigational therapies that might be recommended based on assays results.
Results and Genomic Findings Summary
Here, a member of the clinical genomic interpretation team highlights key findings of the genomic report. Additionally, if a mutation is annotated as either FDA-approved or an investigational biomarker it is highlighted.
Sample Information and Quality
This section outlines key quality control metrics ensuring each component of the clinical assay passing reporting standards as defined in the SOPs.
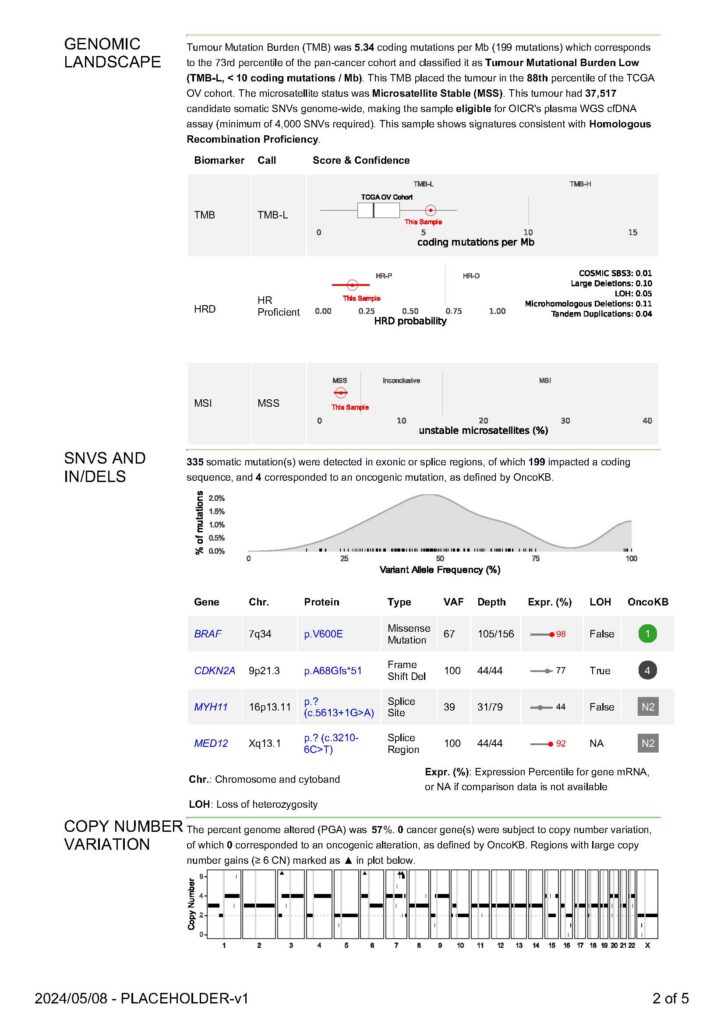
Genomic Landscape
The goal of this section is the situate broad genomic findings such as tumor mutation burden or fraction genome altered against large cohorts of samples, both pan-cancer and cancer-specific where applicable.
Oncogenic Small Mutations and Indels
The analysis pipeline identifies two classes of mutations: (1) single nucleotide variants (SNVs) and (2) indels. Paired sample mutation calling is performed on tumor samples and their respective matched normal controls. Filtering is performed to remove low quality sequence data, sources of sequencing artifacts, and germline results. Mutations which are annotated by OncoKB as Oncogenic or Likely Oncogenic are included in this section.
Supplemental Gene Information
This section contains the chromosomal location and gene description as described by OncoKB. https://www.oncokb.org/cancerGenes
Oncogenic Somatic Copy Number Variation
The analysis pipeline identifies two classes of mutations: (1) Amplifications and (2) Homozygous deletions. Paired sample copy calling is performed on tumor samples and their respective matched normal controls. Filtering is performed to remove low quality sequence data, sources of sequencing artifacts, and germline results. Mutations which are annotated by OncoKB as Oncogenic or Likely Oncogenic are included in this section. An amplification is a mutation that increases the copy number of a specific DNA segment in a cancer cell. A normal diploid genome contains two DNA copies, while amplification increases the DNA copy number. Conversely a homozygous deletion is when the two DNA copies are lost.

Structural Variants and Fusions
The analysis pipeline identifies fusion variants with support from structural variants. A gene fusion is when the transcripts of two genes are joined creating a new gene product.
Assay Description
This section provides further information which may aid in the interpretation of the clinical report. This includes the current versions for the bioinformatic tools utilized in the pipeline, the limit of detection for different mutation types, and key definitions such as the OncoKB levels.



