Enzymatic Methyl-seq (EM-seq)
EM-Seq relies on the NEBNext Enzymatic Methyl-seq kit to enzymatically convert unmethylated cytosines to uracil, preserving the methylated cytosine as cytosine. During PCR, uracil gets converted to thyme. As a result, unmethylated cytosines are sequenced as thyme and methylated cytosines are sequenced as cytosine.
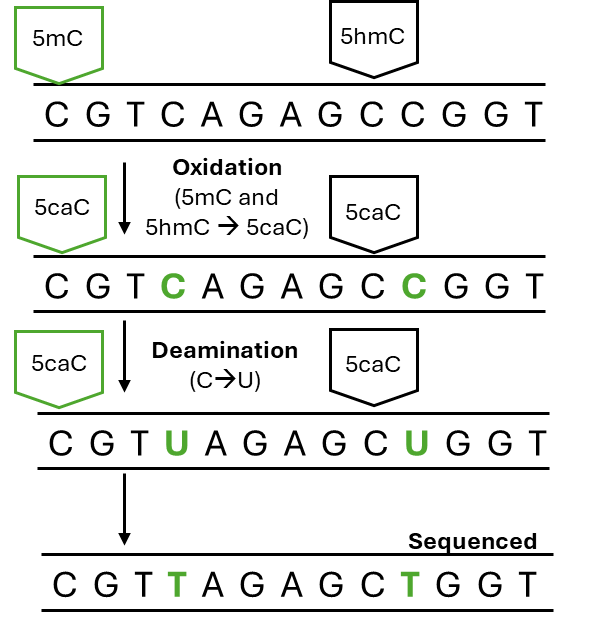
This assays allows for the identification of methylated regions at the single nucleotide resolution with sample input as little as 10 ng gDNA (please note that the identification of 5-methylcytosine (5mC) and 5-hydromethylcytosine (5hmC) are performed in separate assays, Em-Seq and E5hmC-Seq, respectively). You also have the flexibility to sequence the genome and methylome at different depths (Em-Seq and shallow WGS Combo).
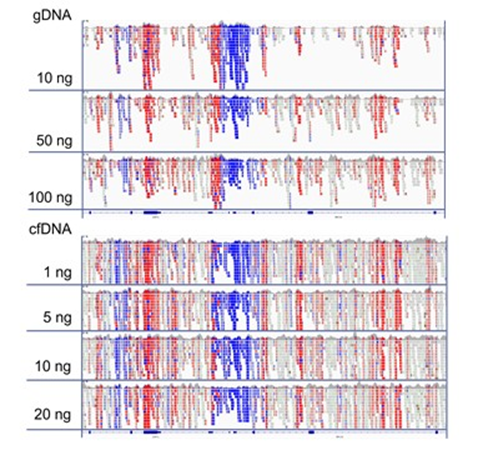
IGV Display of the region around ZAR1L and BRCA2 promoter region.
Blue = Hypo-methylated, Red = Hyper-methylated
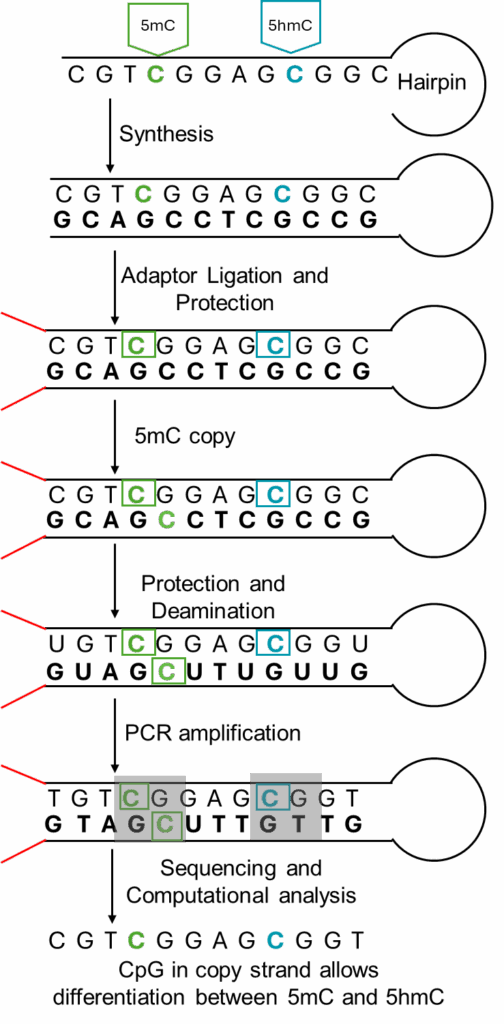
Biomodal Duet Multiomic solution evoC
The Biomodal Duet Multiomic solution evoc (6-letter) technology allows researchers to obtain both genome and methylome information with a single library preparation, while also allowing differentiation between 5mC in 5hmC in a single assay. This technology is especially ideal for samples with limited DNA amount.
This assays utilizes hairpin loops and a DNA methyltransferase in addition to the traditionally used DNA modifying enzymes to “copy” the 5mC patterns onto the copied complimentary strand, ultimately allowing differentiation between 5mC and 5hmC.
Biomodal’s 6-letter’s assay demonstrated similar global methylation detection to Em-Seq and EhmC-Seq across different input type and amounts.
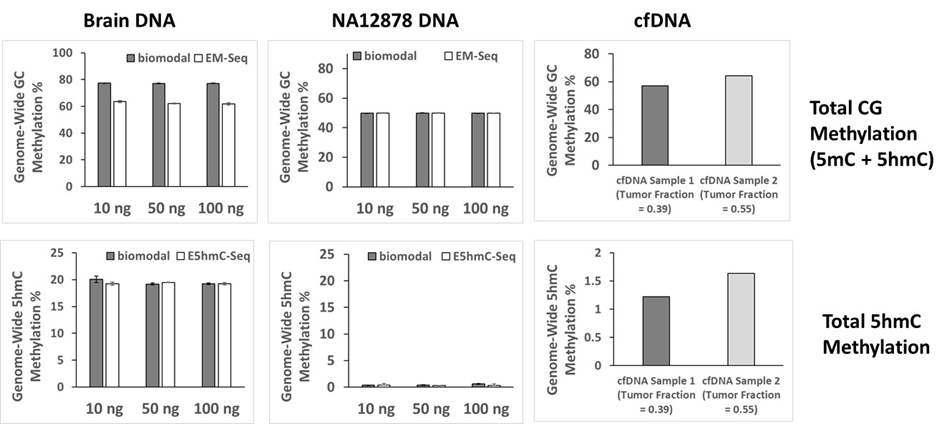
Comparison performed using the Illumina Nova Seq 6000 platform.
Tags:
Posted on:
